 Dear Visitor, this website is dedicated to my results done in cooperation with various international projects. In my research, I use state-of-the-art interdisciplinary approaches for biological problems solving (pre-clinical drug-discovery).
Dear Visitor, this website is dedicated to my results done in cooperation with various international projects. In my research, I use state-of-the-art interdisciplinary approaches for biological problems solving (pre-clinical drug-discovery).
The most common techniques include: sequence alignment, molecular modeling, molecular docking and virtual screening, molecular dynamics simulations of biological macromolecules in silico.
I have completed my PhD in Molecular Biology (2017) at the Protein Engineering and Bioinformatics Department, Institute of Molecular Biology and Genetics, National Academy of Sciences of Ukraine (https://imbg.org.ua/en/). I had Postdoctoral Research (2019) in Bioinformatics and High-Performance Computing Research Group, Catholic University of Murcia (UCAM, South East Spain, https://www.ucam.edu/). From 2022 to 2o23 I worked as Postdoctoral Research Fellow at the Laboratory of Drug Discovery, Design, and Optimization For Novel Therapeutics, Department of Neuroscience, Department of Computational Biology (Div of Health Sciences), Mayo Clinic, Florida, USA. (https://www.mayoclinic.org/). Since 2024 I have started a new role as Chief Scientific Officer (CSO) at Pauling AI, USA (https://www.pauling.ai).
Via oral and poster presentations I participated in more than 30 International Conferences, Congresses and Workshops in EU countries, supported by personal Travel Grants and Certificates from FEBS/IUBMB/EMBO, EBSA, iNEXT (HORIZON 2020), ENABLE (Biotechnology Business Institute), EGI-Engage (HORIZON 2020), PRACE/Cineca, HPC-Europa3 (Horizon 2020), eSSENCE, COMBIOM (7th EU Framework Program) and WIPO, among others. In addition to numerous Awards for the best presentations, I was awarded a four-year Fellowship as one of the best PhD students at the National Academy of Sciences of Ukraine (2012-2014, 2016-2018), Fellowship of President of Ukraine (2018-2019), Kyiv City Mayor Prize (2018), Shevchenko Scientific Society, Inc Award (New York, USA, 2018) and State Prize of President of Ukraine for young scientists in 2018.
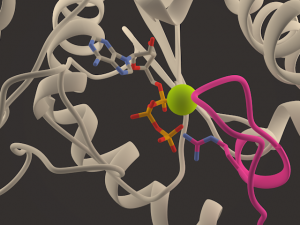
My dissertation combines molecular modeling, molecular docking, MD simulations and analysis of conformational changes in human tyrosyl-tRNA synthetase (HsTyrRS) and its mutant forms, associated with neurodegenerative diseases like Charcot-Marie-Tooth neuropathy (degeneration of peripheral nerve fibers). All automation of HPC computations was supported by in-house developed MolDynGrid Virtual Laboratory (https://moldyngrid.org) as part of the Ukrainian National Grid (https://ung.in.ua/) and European Grid Infrastructure (https://www.egi.eu/). Our research supports the idea that the lack of IL8-like (Interleukin 8) cytokine activity in the full-length HsTyrRS is explained by interactions between N- and C-terminal modules, which block the ELR motif [Savytskyi et al, 2013, J Mol Recognit, 113-120]. We found the new role of connective peptide 1 (CP1) in mammalian tyrosyl-tRNA synthetase. Formation of strong interaction between Lys154 (located in CP1) and γ-phosphate suggests the additional role of CP1 insertion as an important factor for ATP binding. [Kravchuk, Savytskyi et al., 2017, J. Biomol. Struct. Dyn., 2772–2788]. Novel β-sheet formation was observed in the K147-E157 region in G41R and 153-156delVKQV mutant forms of HsTyrRS in the CP1 region of the Rossmann fold. The results support the idea, that defects of the intermolecular interfaces of complexes of mutational forms of HsTyrRS with cognate tRNA(Tyr) and/or translation elongation factor eEF1A may be a common molecular mechanism of Charcot-Marie-Tooth disorder.
As a Research Group Leader, I have developed strong and productive collaborations with students in computational structural biology and drug discovery in silico. It resulted in the graduate studies of Vladyslav Kravchuk at the Institute of Science and Technology (IST) Austria and Master’s study of Valentyn Petrychenko at Georg-August-Universität Göttingen, Germany (2017). My supervision by junior researcher Caleb Weber in cancer and neurological projects led him to be promoted on Special Project Associate II (SPA II) position at Mayo Clinic (2023).
I have authored more than 40 publications (12 papers in international journals with an impact factor of up to 17). I have participated in 7 local and 7 international research projects related to Computational Structural Biology, High-Performance Computing and Grid-infrastructures.
You can find my complete CV at https://alstudio.org/savytskyi/about-oleksandr/cv/ in more detail.
Best Regards,
Oleksandr V. Savytskyi. (updated 13.06.2024)